Power spectra computation in HEALPIX pixellisation#
Introduction#
This tutorial shows how to compute the power spectra of spin 0 and 2 fields. We will use the HEALPIX
pixellisation to pass through the different steps of generation. If you are interested on doing the
same thing with CAR pixellisation, you can have a look at this file.
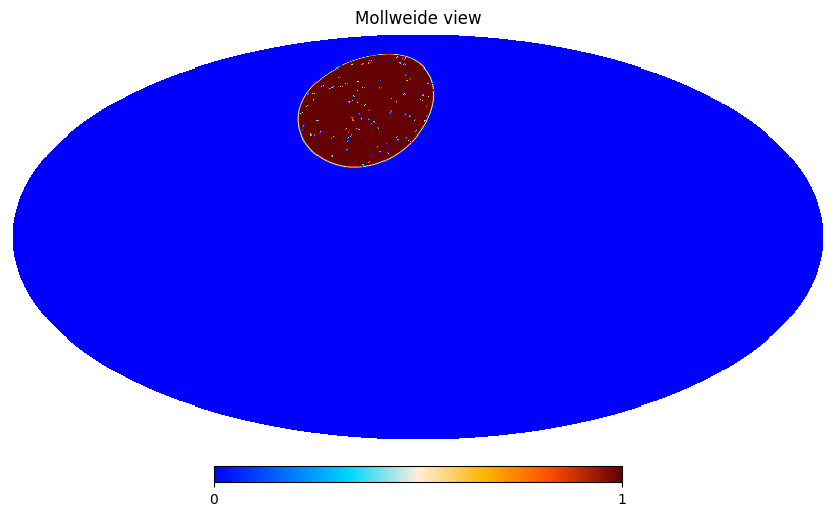
The HEALPIX survey mask will be a disk of radius 25 degree centered on longitude 30 degree and latitude 50 degree, It will have a resolution nside=1024.
We simulate 2 splits with 5 µK.arcmin noise and we also include a point source mask with 100 holes of size 10
arcminutes. We appodize the survey mask with an apodisation of 1 degree and the point source mask
with an apodisation of 0.3 degree. We finally compute the spectra between the 2 splits of data up to
lmax=1000.
Preamble#
Versions used for this tutorial
import healpy as hp
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pixell
import pspy
print(" Numpy :", np.__version__)
print(" Healpy :", hp.__version__)
print("Matplotlib :", mpl.__version__)
print(" pixell :", pixell.__version__)
print(" pspy :", pspy.__version__)
Numpy : 1.26.3
Healpy : 1.16.6
Matplotlib : 3.8.2
pixell : 0.21.1
pspy : 1.7.0+1.gafa4ce9.dirty
Set Planck colormap as default
pixell.colorize.mpl_setdefault("planck")
Generation of the templates, mask and apodisation type#
We start by specifying the HEALPIX parameters for the window function. it will be a disk of radius 25 degree centered on longitude 30 degree and latitude 50 degree, It will have a resolution nside=1024.
lon, lat = 30, 50
radius = 25
nside = 1024
For this example, we will make use of 3 components : Temperature (spin 0) and polarisation Q and U (spin 2)
ncomp = 3
Given the parameters, we can generate the HEALPIX template as follow
from pspy import so_map
template_healpix = so_map.healpix_template(ncomp, nside=nside)
We also define a binary template for the window function: we set pixel inside the disk at 1 and pixel outside at zero
binary_healpix = so_map.healpix_template(ncomp=1, nside=nside)
vec = hp.pixelfunc.ang2vec(lon, lat, lonlat=True)
disc = hp.query_disc(nside, vec, radius=radius * np.pi / 180)
binary_healpix.data[disc] = 1
Generation of spectra#
Generation of simulations#
We first have to compute a set of theoretical power spectra \(C_\ell\) using a Boltzmann solver such as CAMB and we need to install it
since this is a prerequisite of pspy. We can do it within this notebook by executing the following
command
%pip install camb
Requirement already satisfied: camb in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (1.5.3)
Requirement already satisfied: scipy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.11.4)
Requirement already satisfied: sympy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.12)
Requirement already satisfied: packaging in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (23.2)
Requirement already satisfied: numpy<1.28.0,>=1.21.6 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from scipy>=1.0->camb) (1.26.3)
Requirement already satisfied: mpmath>=0.19 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from sympy>=1.0->camb) (1.3.0)
Note: you may need to restart the kernel to use updated packages.
To make sure everything goes well, we can import CAMB and check its version
import camb
print("CAMB version:", camb.__version__)
CAMB version: 1.5.3
Now that CAMB is properly installed, we will produce the \(C_\ell\)s from \(\ell\)min=2 to
\(\ell\)max=104 for the following set of \(\Lambda\)CDM parameters
ℓmin, ℓmax = 2, 10**4
ℓ = np.arange(ℓmin, ℓmax)
cosmo_params = {
"H0": 67.5,
"As": 1e-10 * np.exp(3.044),
"ombh2": 0.02237,
"omch2": 0.1200,
"ns": 0.9649,
"Alens": 1.0,
"tau": 0.0544,
}
pars = camb.set_params(**cosmo_params)
pars.set_for_lmax(ℓmax, lens_potential_accuracy=1)
results = camb.get_results(pars)
powers = results.get_cmb_power_spectra(pars, CMB_unit="muK")
We finally have to write the \(C_\ell\)s into a file to feed the so_map.synfast function
import os
output_dir = "/tmp/tutorial_spectra_spin0and2"
os.makedirs(output_dir, exist_ok=True)
cl_file = os.path.join(output_dir, "cl_camb.dat")
np.savetxt(cl_file, np.hstack([ℓ[:, np.newaxis], powers["total"][ℓmin:ℓmax]]))
Given the CAMB file, we generate a CMB realisation
cmb = template_healpix.synfast(cl_file)
Then, we make 2 splits out of it, each with 5 µK.arcmin rms in temperature and 5xsqrt(2) µK.arcmin in polarisation
nsplits = 2
splits = [cmb.copy() for i in range(nsplits)]
for i in range(nsplits):
noise = so_map.white_noise(cmb, rms_uKarcmin_T=5, rms_uKarcmin_pol=np.sqrt(2) * 5)
splits[i].data += noise.data
We can plot each component T, Q, U for the original CMB realisation
for i, field in enumerate(fields := "TQU"):
hp.mollview(cmb.data[i], title=field)
Generate window#
We then create an apodisation for the survey mask. We use a C1 apodisation with an apodisation size of 1 degree
from pspy import so_window
window = so_window.create_apodization(binary_healpix, apo_type="C1", apo_radius_degree=1)
We also create a point source mask made of 100 holes each with a 10 arcminutes size
mask = so_map.simulate_source_mask(binary_healpix, n_holes=100, hole_radius_arcmin=10)
and we apodize it
mask = so_window.create_apodization(mask, apo_type="C1", apo_radius_degree=0.3)
The window is given by the product of the survey window and the mask window
window.data *= mask.data
hp.mollview(window.data, min=0, max=1)
Compute mode coupling matrix#
For spin 0 and 2 the window need to be a tuple made of two objects: the window used for spin 0 and the one used for spin 2
window = (window, window)
The windows (for spin0 and spin2) are going to couple mode together, we compute a mode coupling
matrix in order to undo this effect given a binning file (format: lmin, lmax, lmean) and a
\(\ell\)max value of 1000
from pspy import pspy_utils
binning_file = os.path.join(output_dir, "binning.dat")
pspy_utils.create_binning_file(bin_size=40, n_bins=100, file_name=binning_file)
from pspy import so_mcm
ℓmax = 1000
mbb_inv, Bbl = so_mcm.mcm_and_bbl_spin0and2(window, binning_file, lmax=ℓmax, type="Dl", niter=0)
Compute alms and bin spectra#
from pspy import sph_tools
alms = [sph_tools.get_alms(split, window, niter=0, lmax=lmax) for split in splits]
We need to specify the order of the spectra to be used by pspy
spectra = ["TT", "TE", "TB", "ET", "BT", "EE", "EB", "BE", "BB"]
and we finally build a dictionary of cross split spectra
Db_dict = {}
from itertools import combinations_with_replacement as cwr
for (i1, alm1), (i2, alm2) in cwr(enumerate(alms), 2):
from pspy import so_spectra
ℓ, ps = so_spectra.get_spectra(alm1, alm2, spectra=spectra)
lb, Db = so_spectra.bin_spectra(
ℓ, ps, binning_file, ℓmax, type="Dl", mbb_inv=mbb_inv, spectra=spectra
)
Db_dict.update({f"split{i1}xsplit{i2}": Db})
To compare with the input \(C_\ell\), we also compute the binned theory spectra
from pspy import pspy_utils
ℓ, ps_theory = pspy_utils.ps_lensed_theory_to_dict(cl_file, "Dl", lmax=ℓmax)
ps_theory_b = so_mcm.apply_Bbl(Bbl, ps_theory, spectra=spectra)
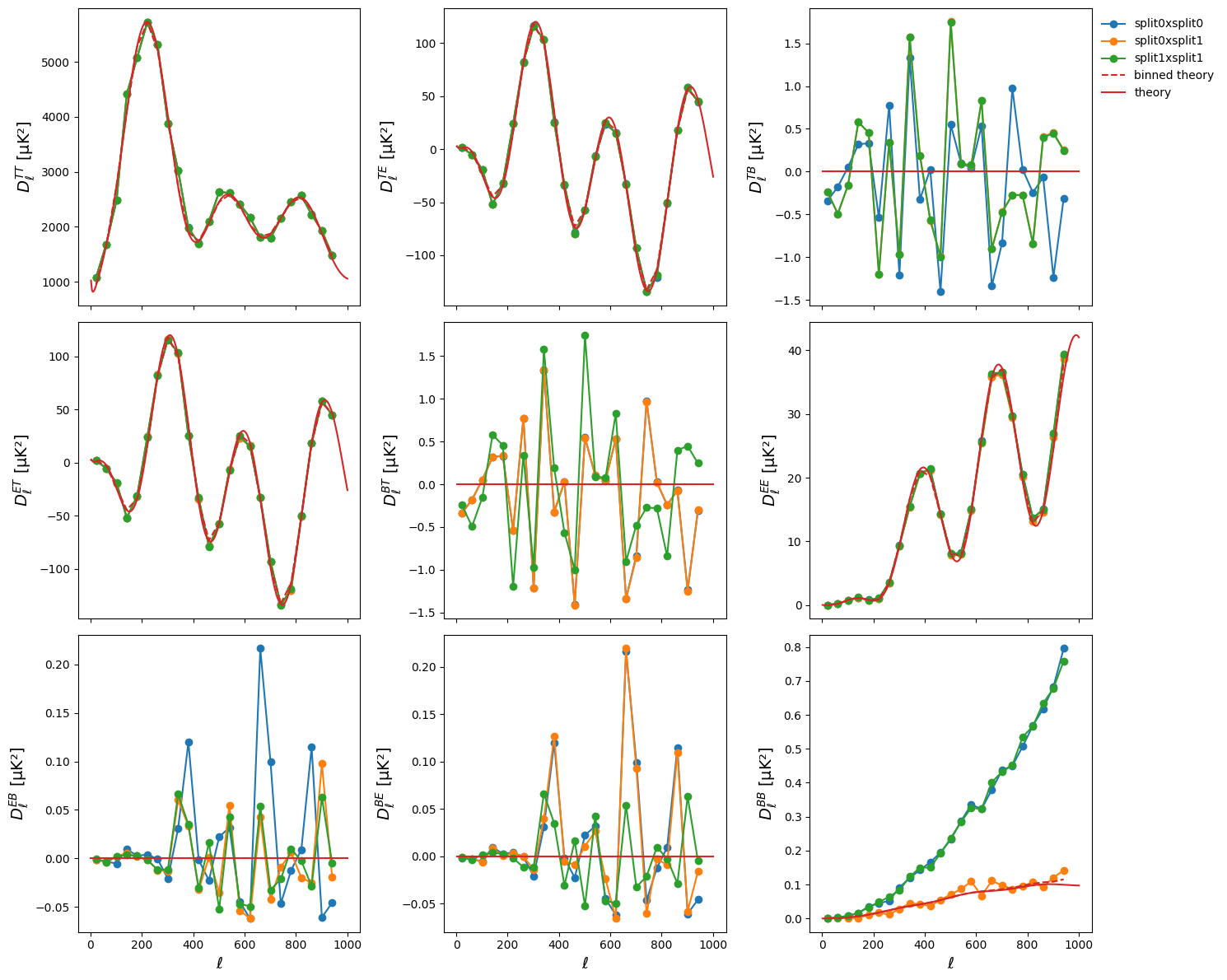
and we finally plot all the results
fig, axes = plt.subplots(3, 3, figsize=(15, 12), sharex=True)
for ax, spec in zip(axes.flatten(), spectra):
for k, v in Db_dict.items():
ax.plot(lb, v[spec], "-o", label=k)
ax.plot(lb, ps_theory_b[spec], "--", color="tab:red", label="binned theory")
ax.plot(l, ps_theory[spec], color="tab:red", label="theory")
ax.set_ylabel(r"$D^{%s}_{\ell}$ [µK²]" % spec, fontsize=14)
for ax in axes[-1]:
ax.set_xlabel(r"$\ell$", fontsize=14)
axes[0, -1].legend(loc="upper left", bbox_to_anchor=(1, 1))
fig.tight_layout()