HEALPIX projection onto CAR pixellisation#
Introduction#
This is a tutorial of generation of simulations and projection. We first specify two templates, one
in equatorial coordinates with CAR pixellisation and one in equatorial coordinates with HEALPIX
pixellisation. We generate alms from a CAMB lensed power spectrum file and use them to generate a
random CMB realisation in both templates. We then project the HEALPIX simulation into the CAR template and plot both the
native CAR simulation and the projected HEALPIX simulation. We chose a low resolution nside to
emphasize the effect of resolution.
Preamble#
Print versions used
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pixell
import pspy
print(" Numpy :", np.__version__)
print("Matplotlib :", mpl.__version__)
print(" pixell :", pixell.__version__)
print(" pspy :", pspy.__version__)
Numpy : 1.26.3
Matplotlib : 3.8.2
pixell : 0.21.1
pspy : 1.7.0+1.gafa4ce9.dirty
Set Planck colormap as default
pixell.colorize.mpl_setdefault("planck")
Generation of the templates#
The CAR template will go from right ascension ra0 to ra1 and from declination dec0 to dec1 (all in
degrees). It will have a resolution of 1 arcminute and it allows 3 components (stokes parameter in
the case of CMB anisotropies).
ra0, ra1 = -5, 5
dec0, dec1 = -5, 5
res = 1
ncomp = 3
from pspy import so_map
template_car = so_map.car_template(ncomp, ra0, ra1, dec0, dec1, res)
We also generate an HEALPIX template for which we choose nside=256 so that the resolution of HEALPIX
is much smaller
template_healpix = so_map.healpix_template(ncomp, nside=256, coordinate="equ")
Read power spectrum and alm generation#
We first have to compute the power spectra \(C_\ell\)s using a Boltzmann solver such as CAMB and we need to install it
since this is a prerequisite of pspy. We can do it within this notebook by executing the following
command
%pip install camb
Requirement already satisfied: camb in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (1.5.3)
Requirement already satisfied: scipy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.11.4)
Requirement already satisfied: sympy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.12)
Requirement already satisfied: packaging in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (23.2)
Requirement already satisfied: numpy<1.28.0,>=1.21.6 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from scipy>=1.0->camb) (1.26.3)
Requirement already satisfied: mpmath>=0.19 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from sympy>=1.0->camb) (1.3.0)
Note: you may need to restart the kernel to use updated packages.
To make sure everything goes well, we can import CAMB and check its version
import camb
print("CAMB version:", camb.__version__)
CAMB version: 1.5.3
Now that CAMB is properly installed, we will produce \(C_\ell\) data from \(\ell\)min=2 to
\(\ell\)max=104 for the following set of \(\Lambda\)CDM parameters
ℓmin, ℓmax = 2, 10**4
ℓ = np.arange(ℓmin, ℓmax)
cosmo_params = {
"H0": 67.5,
"As": 1e-10 * np.exp(3.044),
"ombh2": 0.02237,
"omch2": 0.1200,
"ns": 0.9649,
"Alens": 1.0,
"tau": 0.0544,
}
pars = camb.set_params(**cosmo_params)
pars.set_for_lmax(ℓmax, lens_potential_accuracy=1)
results = camb.get_results(pars)
powers = results.get_cmb_power_spectra(pars, CMB_unit="muK")
We finally have to write \(C_\ell\) into a file to read back using the pixell.powspec function
import os
output_dir = "/tmp/tutorial_projection"
os.makedirs(output_dir, exist_ok=True)
cl_file = os.path.join(output_dir, "cl_camb.dat")
np.savetxt(cl_file, np.hstack([ℓ[:, np.newaxis], powers["total"][ℓmin:ℓmax]]))
from pixell import powspec
ps = powspec.read_spectrum(cl_file)[:ncomp, :ncomp]
and generate alms from the power spectrum up to lmax = 5000
from pixell import curvedsky
ℓmax = 5000
alms = curvedsky.rand_alm(ps, lmax=lmax)
Computation of stokes parameters#
We compute the stokes parameters from the alms in both templates
from pspy import sph_tools
map_healpix = sph_tools.alm2map(alms, template_healpix)
map_car = sph_tools.alm2map(alms, template_car)
and we project the HEALPIX map into the CAR template
map_healpix_proj = so_map.healpix2car(map_healpix, map_car, lmax=lmax)
WARNING: your lmax is too large, setting it to 3*nside-1 now
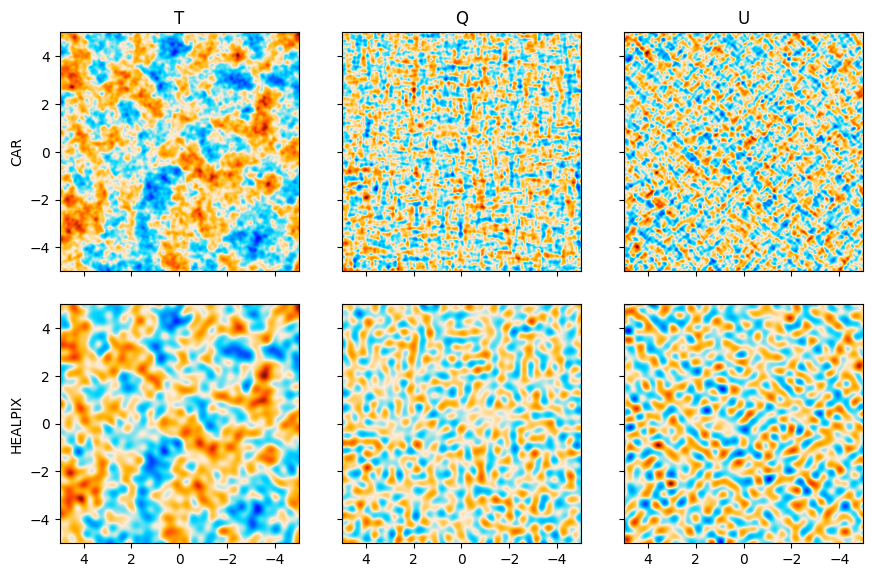
Showing maps#
We plot both the native CAR map and the HEALPIX projected to CAR map. They contain the same CMB but
have different resolutions.
fig, axes = plt.subplots(2, 3, figsize=(9, 6), sharex=True, sharey=True)
kwargs = dict(extent=[ra1, ra0, dec0, dec1], origin="lower")
for i, field in enumerate(fields := "TQU"):
kwargs["vmin"] = np.min([map_car.data[i], map_healpix_proj.data[i]])
kwargs["vmax"] = np.max([map_car.data[i], map_healpix_proj.data[i]])
axes[0, i].imshow(map_car.data[i], **kwargs)
axes[1, i].imshow(map_healpix_proj.data[i], **kwargs)
axes[0, i].set_title(field)
axes[0, 0].set_ylabel("CAR")
axes[1, 0].set_ylabel("HEALPIX")
fig.tight_layout()
We can also use the plot function from pspy.so_map and set the output path to get individual images
for each component T, Q, U.
map_car.plot(file_name=os.path.join(output_dir, "map_car"))
map_healpix_proj.plot(file_name=os.path.join(output_dir, "map_healpix"))
