Compute spectra for standard and pure B modes#
Introduction#
This tutorial illustrates the spectra computation for standard and pure B modes. We will only use
the HEALPIX pixellisation to pass through the different steps of generation.
The HEALPIX survey mask is a disk centered on longitude 30° and latitude 50° with a radius of 25
radians. The nside value is set to 512 for this tutorial to reduce computation time.
Preamble#
Versions used for this tutorial
import healpy as hp
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pixell
import pspy
print(" Numpy :", np.__version__)
print(" Healpy :", hp.__version__)
print("Matplotlib :", mpl.__version__)
print(" pixell :", pixell.__version__)
print(" pspy :", pspy.__version__)
Numpy : 1.26.3
Healpy : 1.16.6
Matplotlib : 3.8.2
pixell : 0.21.1
pspy : 1.7.0+1.gafa4ce9.dirty
Set Planck colormap as default
pixell.colorize.mpl_setdefault("planck")
Generation of the templates, mask and apodisation type#
We start by specifying the HEALPIX survey parameters namely longitude, latitude and patch size. The
nside value is set to 512.
lon, lat = 30, 50
radius = 25
nside = 512
Given the nside value, we can set the \(\ell\)max value
ℓmax = 3 * nside - 1
For this example, we will make use of 3 components : Temperature (spin 0) and polarisation Q and U (spin 2)
ncomp = 3
Given the parameters, we can generate the HEALPIX template as follow
from pspy import so_map
template = so_map.healpix_template(ncomp, nside)
We also define the binary template for the window function pixels
binary = so_map.healpix_template(ncomp=1, nside=nside)
vec = hp.pixelfunc.ang2vec(lon, lat, lonlat=True)
disc = hp.query_disc(nside, vec, radius=radius * np.pi / 180)
binary.data[disc] = 1
Generation of spectra#
Generate window#
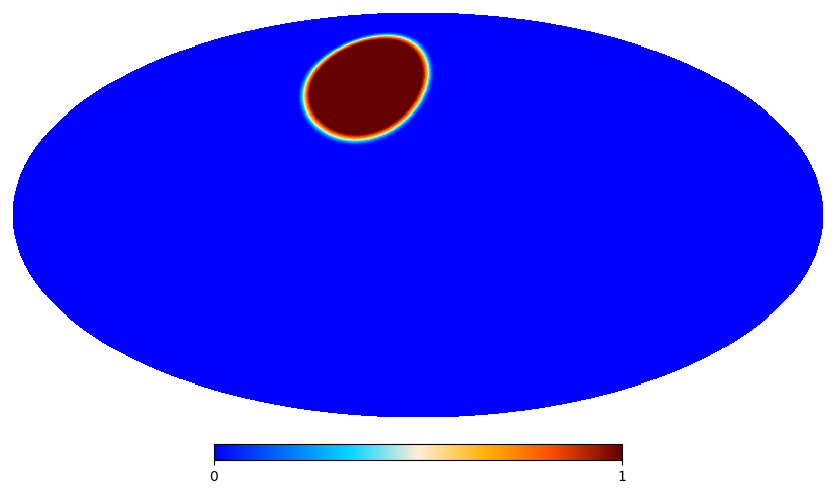
We then create an apodisation for the survey mask. We use a C1 apodisation with an apodisation size of 5 degrees
from pspy import so_window
window = so_window.create_apodization(binary, apo_type="C1", apo_radius_degree=5)
hp.mollview(window.data, title=None)
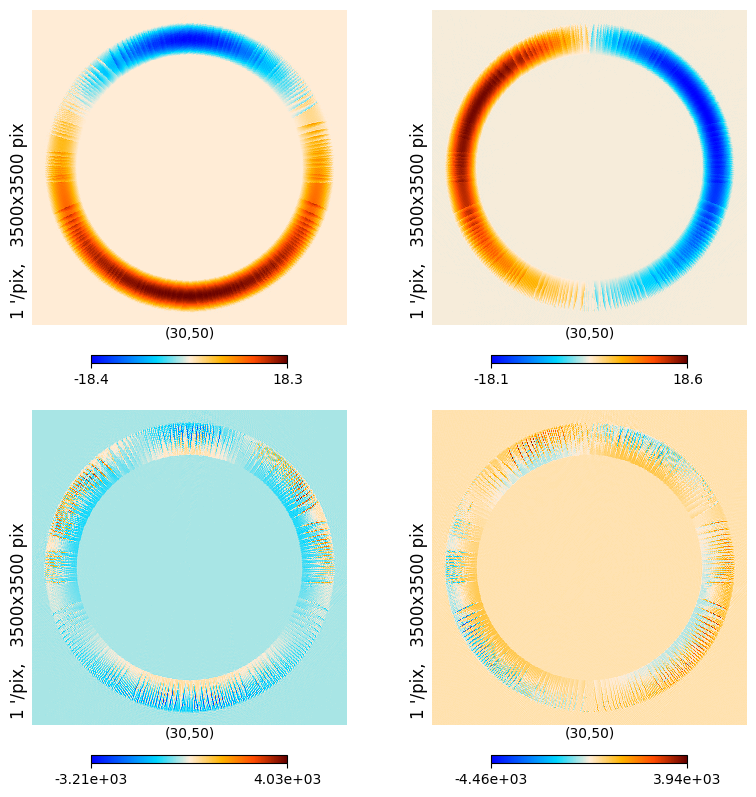
We can also have a look to the corresponding spin 1 and spin 2 window functions
niter = 3
w1_plus, w1_minus, w2_plus, w2_minus = so_window.get_spinned_windows(window, lmax=ℓmax, niter=niter)
plt.figure(figsize=(8, 8))
kwargs = {"rot": (lon, lat, 0), "xsize": 3500, "reso": 1, "title": None}
hp.gnomview(w1_plus.data, **kwargs, sub=(2, 2, 1))
hp.gnomview(w1_minus.data, **kwargs, sub=(2, 2, 2))
hp.gnomview(w2_plus.data, **kwargs, sub=(2, 2, 3))
hp.gnomview(w2_minus.data, **kwargs, sub=(2, 2, 4))
Binning file#
We create a binning file with the following format : lmin, lmax, lmean
import os
from pspy import pspy_utils
output_dir = "/tmp/tutorial_purebb"
os.makedirs(output_dir, exist_ok=True)
binning_file = os.path.join(output_dir, "binning.dat")
pspy_utils.create_binning_file(bin_size=50, n_bins=300, file_name=binning_file)
Compute mode coupling matrix#
For spin 0 and 2 the window need to be a tuple made of two objects: the window used for spin 0 and the one used for spin 2
window_tuple = (window, window)
The windows (for spin0 and spin2) are going to couple mode together, we compute a mode coupling
matrix in order to undo this effect given the binning file. We do it for both calculations i.e.
standard and pure B mode
from pspy import so_mcm
kwargs = dict(win1=window_tuple, binning_file=binning_file, lmax=ℓmax, niter=niter, type="Cl")
print("Computing standard mode coupling matrix")
mbb_inv, Bbl = so_mcm.mcm_and_bbl_spin0and2(**kwargs)
print("Computing pure mode coupling matrix")
mbb_inv_pure, Bbl_pure = so_mcm.mcm_and_bbl_spin0and2(**kwargs, pure=True)
Show code cell output
Computing standard mode coupling matrix
Computing pure mode coupling matrix
Generation of ΛCDM power spectra#
We first have to compute \(C_\ell\) data using a cosmology code such as CAMB and we need to install it
since this is not a prerequisite of pspy. We can do it within this notebook by executing the
following command
%pip install camb
Requirement already satisfied: camb in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (1.5.3)
Requirement already satisfied: scipy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.11.4)
Requirement already satisfied: sympy>=1.0 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (1.12)
Requirement already satisfied: packaging in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from camb) (23.2)
Requirement already satisfied: numpy<1.28.0,>=1.21.6 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from scipy>=1.0->camb) (1.26.3)
Requirement already satisfied: mpmath>=0.19 in /home/garrido/Workdir/cmb/development/pspy/pyenv/lib/python3.11/site-packages (from sympy>=1.0->camb) (1.3.0)
Note: you may need to restart the kernel to use updated packages.
To make sure everything goes well, we can import CAMB and check its version
import camb
print("CAMB version:", camb.__version__)
CAMB version: 1.5.3
Now that CAMB is properly installed, we will produce the \(C_\ell\)s from \(\ell\)min=2 to
\(\ell\)max=104 for the following set of \(\Lambda\)CDM parameters
ℓ = np.arange(2, 10_000)
cosmo_params = {
"H0": 67.5,
"As": 1e-10 * np.exp(3.044),
"ombh2": 0.02237,
"omch2": 0.1200,
"ns": 0.9649,
"Alens": 1.0,
"tau": 0.0544,
}
pars = camb.set_params(**cosmo_params)
pars.set_for_lmax(ℓ[-1], lens_potential_accuracy=1)
results = camb.get_results(pars)
powers = results.get_cmb_power_spectra(pars, CMB_unit="muK")
We finally have to write \(C_\ell\) into a file to feed the so_map.synfast function
cl_file = os.path.join(output_dir, "cl_camb.dat")
np.savetxt(cl_file, np.hstack([ℓ[:, np.newaxis], powers["total"][ℓ]]))
Running simulations#
Given the parameters and data above, we will now simulate n_sims simulations to check for mean and
variance of BB spectrum. For illustrative purpose, we will only run 10 simulations (~ few minutes)
but for reasonable comparisons, you should increase this number to few tens of simulations.
We will do it for both calculations (standard and pure) and finally we will graphically compare results
We first need to specify the order of the spectra to be used by pspy although only BB spectrum will
be used
spectra = ["TT", "TE", "TB", "ET", "BT", "EE", "EB", "BE", "BB"]
and we define a dictionnary of methods regarding the calculation type for B mode spectrum
from pspy import sph_tools
methods = {
"standard": dict(alm=sph_tools.get_alms, mbb=mbb_inv),
"pure": dict(alm=sph_tools.get_pure_alms, mbb=mbb_inv_pure),
}
from pspy import so_spectra
from tqdm.auto import tqdm
for _ in tqdm(range(n_sims := 10)):
cmb = template.synfast(cl_file)
for k, v in methods.items():
get_alm = v.get("alm")
alm = get_alm(cmb, window_tuple, niter, ℓmax)
ℓ, ps = so_spectra.get_spectra(alm, spectra=spectra)
ℓb, ps_dict = so_spectra.bin_spectra(
ℓ, ps, binning_file, ℓmax, type="Cl", mbb_inv=v.get("mbb"), spectra=spectra
)
v.setdefault("bb", []).append(ps_dict["BB"])
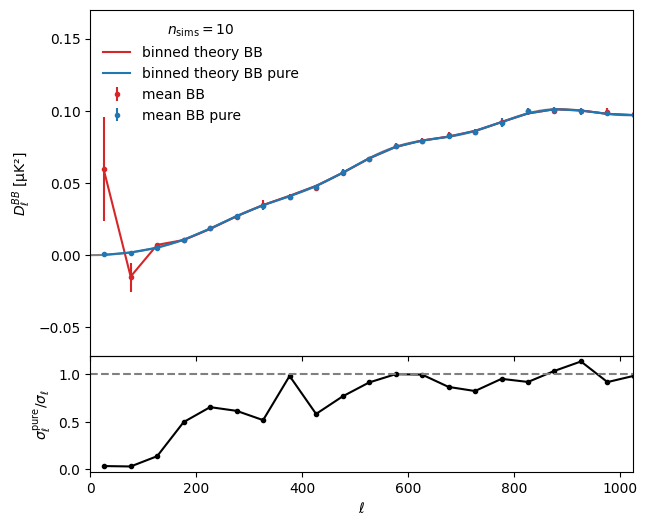
Let’s plot the mean results against the theory value for BB spectrum
for k, v in methods.items():
v["mean"] = np.mean(v.get("bb"), axis=0)
v["std"] = np.std(v.get("bb"), axis=0)
ℓ_th, ps_theory = pspy_utils.ps_lensed_theory_to_dict(cl_file, output_type="Cl", lmax=ℓmax)
ps_theory_b = so_mcm.apply_Bbl(Bbl, ps_theory, spectra=spectra)
ps_theory_b_pure = so_mcm.apply_Bbl(Bbl_pure, ps_theory, spectra=spectra)
fac = ℓb * (ℓb + 1) / (2 * np.pi)
facth = ℓ_th * (ℓ_th + 1) / (2 * np.pi)
fig = plt.figure(figsize=(7, 6))
grid = plt.GridSpec(4, 1, hspace=0, wspace=0)
main = fig.add_subplot(grid[:3], xticklabels=[], xlim=(0, 2 * nside))
main.plot(ℓ_th[:ℓmax], ps_theory["BB"][:ℓmax] * facth[:ℓmax], color="grey")
main.errorbar(ℓb, ps_theory_b["BB"] * fac, color="tab:red", label="binned theory BB")
main.errorbar(ℓb, ps_theory_b_pure["BB"] * fac, color="tab:blue", label="binned theory BB pure")
main.errorbar(
ℓb,
methods.get("standard").get("mean") * fac,
methods.get("standard").get("std") * fac,
fmt=".",
color="tab:red",
label="mean BB",
)
main.errorbar(
ℓb,
methods.get("pure").get("mean") * fac,
methods.get("pure").get("std") * fac,
fmt=".",
color="tab:blue",
label="mean BB pure",
)
main.set(ylim=(-0.07, 0.17), ylabel=r"$D^{BB}_{\ell}$ [µK²]")
plt.legend(title=r"$n_{\rm sims}=%s$" % n_sims)
ratio = fig.add_subplot(grid[3], xlim=(0, 2 * nside))
ratio.plot(ℓb, methods.get("pure").get("std") / methods.get("standard").get("std"), ".-k")
ratio.set(ylabel=r"$\sigma^{\rm pure}_\ell/ \sigma_\ell$", xlabel=r"$\ell$")
ratio.axhline(1, color="gray", ls="dashed");